Abstract
Treatment of acute myeloid leukemia remains inadequate, largely due to our limited understanding of therapy resistance and lack of effective therapeutic targets. Recently, we and others have used functional genomic and proteomic methods to elucidate the mechanisms of therapy resistance. These studies have revealed the existence of privileged populations of AML cells with distinct signaling properties. AML is known to have leukemia stem cells (LSCs), which exhibit self-renewal and cell-cycle quiescence, and contribute to chemotherapy resistance. However, their prospective isolation has been hindered by their variable immunophenotypes, constituting an important gap to understanding their biology, as a prelude to developing improved therapies.
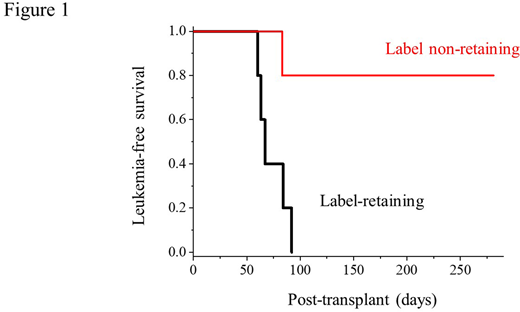
Here, we used chemical protein labeling to prospectively isolate AML quiescent and stem cells from primary patient leukemias, based on proteome production and renewal. First, we developed optimized protein-labeling conditions to preserve stem cell function, as demonstrated by transplantation of labeled mouse hematopoietic stem cells into lethally irradiated recipients. We applied this method to prospectively isolate primary patient AML quiescent cells upon transplanting labeled specimens into immunodeficient NSG mice. These studies showed that label-retaining, but not their label non-retaining populations, are comprised by AML stem cells, as evidenced by serial transplantation and limiting dilution (Figure 1). Using genome sequencing of label-retaining and non-retaining populations, we confirmed the absence of genetic mutations associated with AML proteome quiescence, consistent with their epigenetic basis. To confirm this epigenetic mechanism directly, we performed re-labeling experiments, observing reversible development of quiescent stem cells from their proliferating counterparts. To define the epigenetic mechanisms of AML quiescence, we used assays for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) and mRNA sequencing (RNA-seq) of label-retaining versus non-retaining populations from three different therapy-resistant MLL-, MOZ-, and NUP98-rearranged patient leukemias. Gene set enrichment analysis demonstrated significant association with reported gene expression programs associated with hematopoietic and AML stem cell function, and FACS analysis showed the presence of some but not all reported stem cell surface markers, including CD34, CD38, CD117, CD90, CD45RA, and CD123. Remarkably, in spite of the biological differences among these subtypes, we observed a common set of genes and differentially accessible chromatin loci in AML quiescent as compared to proliferating cells. Motif analysis revealed that differentially accessible chromatin in the label-retaining cell population was predominantly comprised by the E26 transformation-specific (ETS) family transcription factor (TF)-binding motifs. For example, the ETS-related gene (ERG)-binding motif covered more than 50% of differentially accessible chromatin in the label-retaining cell population (p = 10e-7). Consistently, label-retaining quiescent cells exhibited significant induction of gene expression programs associated with ETS transcription factor function.
In summary, this work presents a functional approach for the prospective isolation of primary patient AML quiescent cells. Human AML quiescent cells constitute a distinct population with leukemia-initiating and self-renewal capacity, as well as epigenetic plasticity and reversible induction in diverse genetic disease subtypes. We anticipate that the reported gene expression and chromatin profiles, including those controlled by ETS transcription factors, should define their control mechanisms and targets for improved therapy.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal